Browser
Protein data
function: electron transporterexperiment: ELECTRON MICROSCOPY
resolution: 3.24 Å
origomeric count: 79
axial ligand #1: HIS
chainID: QC,resSeq: 83,
Coordination distance[Å]: 1.978
molecule: Cytochrome b
Organism: Sus scrofa
axial ligand #2: HIS
chainID: QC,resSeq: 182,
Coordination distance[Å]: 1.952
molecule: same as ligand #1
List of other hemes in pdb:8zo8
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
8zo8-C1-602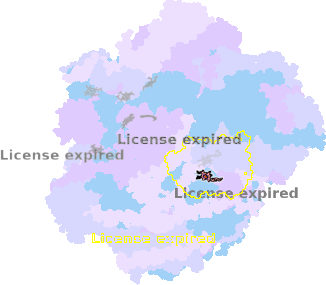 |
sad. +0.01 ruf. +0.16 dom. +0.02 bre. -0.39 |
HIS | chainID: C1, resSeq: 61, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 79 pocket vol.: 645.0 Å3 d(Fe-oop): 0.003 Å |
| HIS | chainID: C1, resSeq: 378, molecule: Cytochrome c oxidase subunit 1 |
|||
8zo8-C1-603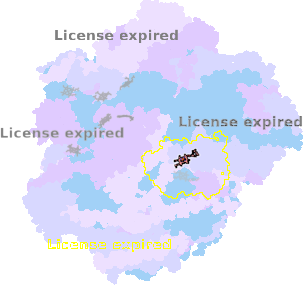 |
sad. +0.02 ruf. +0.07 dom. +0.02 bre. -0.39 |
HIS | chainID: C1, resSeq: 376, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 79 pocket vol.: 649.0 Å3 d(Fe-oop): 0.003 Å |
| Not assigned. | ||||
8zo8-QC-403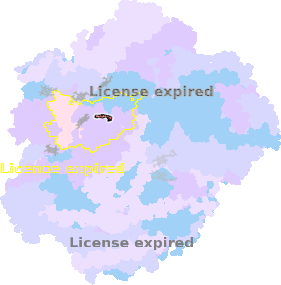 |
sad. -0.09 ruf. -0.99 dom. +0.10 bre. +0.11 |
HIS | chainID: QC, resSeq: 97, molecule: Cytochrome b |
electron transporter oligomeric count: 79 pocket vol.: 432.0 Å3 d(Fe-oop): 0.037 Å |
| HIS | chainID: QC, resSeq: 196, molecule: Cytochrome b |
|||
8zo8-QD-401 |
sad. -0.00 ruf. -0.20 dom. -0.22 bre. -0.03 |
HIS | chainID: QD, resSeq: 126, molecule: Cytochrome c domain-containing protein |
electron transporter oligomeric count: 79 pocket vol.: 416.0 Å3 d(Fe-oop): 0.084 Å |
| MET | chainID: QD, resSeq: 245, molecule: Cytochrome c domain-containing protein |
|||
8zo8-Qc-402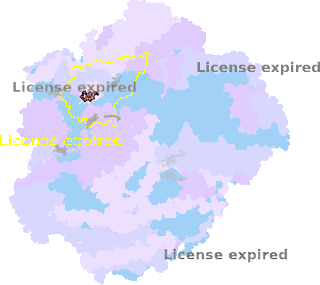 |
sad. +0.00 ruf. -0.26 dom. -0.05 bre. +0.04 |
HIS | chainID: Qc, resSeq: 83, molecule: Cytochrome b |
electron transporter oligomeric count: 79 pocket vol.: 415.0 Å3 d(Fe-oop): 0.016 Å |
| HIS | chainID: Qc, resSeq: 182, molecule: Cytochrome b |
|||
8zo8-Qc-403 |
sad. -0.09 ruf. -0.48 dom. +0.15 bre. +0.05 |
HIS | chainID: Qc, resSeq: 97, molecule: Cytochrome b |
electron transporter oligomeric count: 79 pocket vol.: 416.0 Å3 d(Fe-oop): 0.052 Å |
| HIS | chainID: Qc, resSeq: 196, molecule: Cytochrome b |
|||
8zo8-Qd-402 |
sad. -0.02 ruf. -0.35 dom. -0.04 bre. -0.04 |
HIS | chainID: Qd, resSeq: 126, molecule: Cytochrome c domain-containing protein |
electron transporter oligomeric count: 79 pocket vol.: 452.0 Å3 d(Fe-oop): 0.016 Å |
| MET | chainID: Qd, resSeq: 245, molecule: Cytochrome c domain-containing protein |
|||
