Browser
Protein data
function: oxidoreductaseexperiment: X-RAY DIFFRACTION
resolution: 1.737 Å
origomeric count: 3
axial ligand #1: HIS
chainID: AAA,resSeq: 58,
Coordination distance[Å]: 1.92
molecule: Octaheme nitrite reductase
Organism: Trichlorobacter ammonificans
axial ligand #2: HIS
chainID: AAA,resSeq: 95,
Coordination distance[Å]: 1.905
molecule: same as ligand #1
List of other hemes in pdb:8rxu
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
8rxu-AAA-601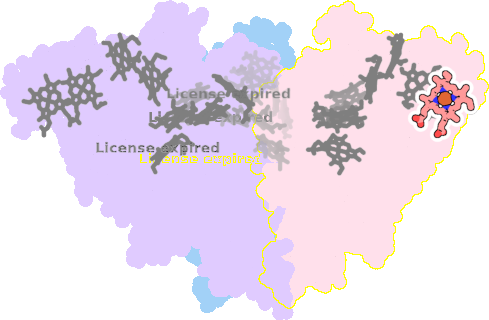 |
sad. +0.04 ruf. -0.73 dom. -0.02 bre. -0.32 |
HIS | chainID: AAA, resSeq: 46, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 546.0 Å3 d(Fe-oop): 0.014 Å |
| HIS | chainID: AAA, resSeq: 73, molecule: Octaheme nitrite reductase |
|||
8rxu-AAA-602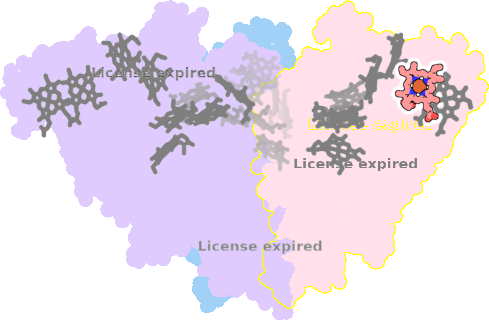 |
sad. +0.03 ruf. -0.57 dom. -0.06 bre. -0.31 |
HIS | chainID: AAA, resSeq: 67, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 368.0 Å3 d(Fe-oop): 0.024 Å |
| HIS | chainID: AAA, resSeq: 250, molecule: Octaheme nitrite reductase |
|||
8rxu-AAA-604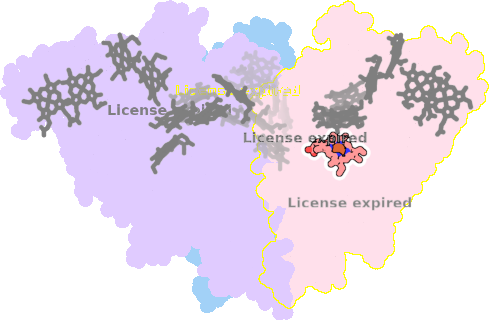 |
sad. +0.20 ruf. -0.75 dom. +0.10 bre. -0.30 |
LYS | chainID: AAA, resSeq: 206, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 394.0 Å3 d(Fe-oop): 0.031 Å |
| PO4 | chainID: AAA, resSeq: 610, molecule: PHOSPHATE ION |
|||
8rxu-AAA-605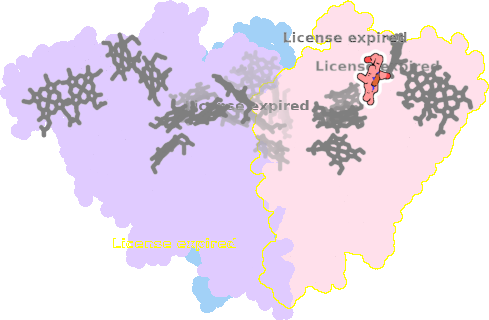 |
sad. +0.53 ruf. -1.03 dom. -0.13 bre. -0.25 |
HIS | chainID: AAA, resSeq: 247, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 462.0 Å3 d(Fe-oop): 0.052 Å |
| HIS | chainID: AAA, resSeq: 407, molecule: Octaheme nitrite reductase |
|||
8rxu-AAA-606 |
sad. -0.26 ruf. -1.23 dom. +0.19 bre. -0.27 |
HIS | chainID: AAA, resSeq: 145, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 425.0 Å3 d(Fe-oop): 0.030 Å |
| HIS | chainID: AAA, resSeq: 309, molecule: Octaheme nitrite reductase |
|||
8rxu-AAA-607 |
sad. +0.70 ruf. -1.19 dom. -0.08 bre. -0.10 |
HIS | chainID: AAA, resSeq: 392, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 364.0 Å3 d(Fe-oop): 0.030 Å |
| HIS | chainID: AAA, resSeq: 500, molecule: Octaheme nitrite reductase |
|||
8rxu-AAA-608 |
sad. -0.15 ruf. -1.61 dom. -0.04 bre. -0.16 |
HIS | chainID: AAA, resSeq: 381, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 481.0 Å3 d(Fe-oop): 0.000 Å |
| HIS | chainID: AAA, resSeq: 424, molecule: Octaheme nitrite reductase |
|||
8rxu-MMM-601 |
sad. +0.03 ruf. -0.90 dom. -0.04 bre. -0.40 |
HIS | chainID: MMM, resSeq: 46, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 536.0 Å3 d(Fe-oop): 0.004 Å |
| HIS | chainID: MMM, resSeq: 73, molecule: Octaheme nitrite reductase |
|||
8rxu-MMM-602 |
sad. -0.07 ruf. -0.29 dom. -0.02 bre. -0.30 |
HIS | chainID: MMM, resSeq: 67, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 352.0 Å3 d(Fe-oop): 0.025 Å |
| HIS | chainID: MMM, resSeq: 250, molecule: Octaheme nitrite reductase |
|||
8rxu-MMM-603 |
sad. +0.20 ruf. -0.16 dom. +0.15 bre. -0.37 |
HIS | chainID: MMM, resSeq: 58, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 524.0 Å3 d(Fe-oop): 0.086 Å |
| HIS | chainID: MMM, resSeq: 95, molecule: Octaheme nitrite reductase |
|||
8rxu-MMM-604 |
sad. +0.32 ruf. -0.53 dom. +0.15 bre. -0.28 |
LYS | chainID: MMM, resSeq: 206, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 394.0 Å3 d(Fe-oop): 0.013 Å |
| PO4 | chainID: MMM, resSeq: 610, molecule: PHOSPHATE ION |
|||
8rxu-MMM-605 |
sad. +0.51 ruf. -0.84 dom. -0.19 bre. -0.35 |
HIS | chainID: MMM, resSeq: 247, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 468.0 Å3 d(Fe-oop): 0.072 Å |
| HIS | chainID: MMM, resSeq: 407, molecule: Octaheme nitrite reductase |
|||
8rxu-MMM-606 |
sad. -0.24 ruf. -0.67 dom. +0.11 bre. -0.27 |
HIS | chainID: MMM, resSeq: 145, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 407.0 Å3 d(Fe-oop): 0.006 Å |
| HIS | chainID: MMM, resSeq: 309, molecule: Octaheme nitrite reductase |
|||
8rxu-MMM-607 |
sad. +0.72 ruf. -0.93 dom. -0.16 bre. -0.14 |
HIS | chainID: MMM, resSeq: 392, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 362.0 Å3 d(Fe-oop): 0.060 Å |
| HIS | chainID: MMM, resSeq: 500, molecule: Octaheme nitrite reductase |
|||
8rxu-MMM-608 |
sad. -0.18 ruf. -1.75 dom. +0.13 bre. -0.15 |
HIS | chainID: MMM, resSeq: 381, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 460.0 Å3 d(Fe-oop): 0.059 Å |
| HIS | chainID: MMM, resSeq: 424, molecule: Octaheme nitrite reductase |
|||
8rxu-YYY-601 |
sad. -0.05 ruf. -0.94 dom. -0.06 bre. -0.33 |
HIS | chainID: YYY, resSeq: 46, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 528.0 Å3 d(Fe-oop): 0.001 Å |
| HIS | chainID: YYY, resSeq: 73, molecule: Octaheme nitrite reductase |
|||
8rxu-YYY-602 |
sad. -0.10 ruf. -0.52 dom. +0.01 bre. -0.37 |
HIS | chainID: YYY, resSeq: 67, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 371.0 Å3 d(Fe-oop): 0.026 Å |
| HIS | chainID: YYY, resSeq: 250, molecule: Octaheme nitrite reductase |
|||
8rxu-YYY-603 |
sad. +0.20 ruf. -0.55 dom. -0.09 bre. -0.48 |
HIS | chainID: YYY, resSeq: 58, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 542.0 Å3 d(Fe-oop): 0.041 Å |
| HIS | chainID: YYY, resSeq: 95, molecule: Octaheme nitrite reductase |
|||
8rxu-YYY-604 |
sad. +0.37 ruf. -0.49 dom. +0.01 bre. -0.36 |
LYS | chainID: YYY, resSeq: 206, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 392.0 Å3 d(Fe-oop): 0.057 Å |
| PO4 | chainID: YYY, resSeq: 610, molecule: PHOSPHATE ION |
|||
8rxu-YYY-605 |
sad. +0.52 ruf. -0.95 dom. -0.08 bre. -0.29 |
HIS | chainID: YYY, resSeq: 247, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 485.0 Å3 d(Fe-oop): 0.002 Å |
| HIS | chainID: YYY, resSeq: 407, molecule: Octaheme nitrite reductase |
|||
8rxu-YYY-606 |
sad. -0.20 ruf. -1.15 dom. +0.05 bre. -0.25 |
HIS | chainID: YYY, resSeq: 145, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 407.0 Å3 d(Fe-oop): 0.040 Å |
| HIS | chainID: YYY, resSeq: 309, molecule: Octaheme nitrite reductase |
|||
8rxu-YYY-607 |
sad. +0.71 ruf. -1.17 dom. -0.18 bre. -0.16 |
HIS | chainID: YYY, resSeq: 392, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 377.0 Å3 d(Fe-oop): 0.040 Å |
| HIS | chainID: YYY, resSeq: 500, molecule: Octaheme nitrite reductase |
|||
8rxu-YYY-608 |
sad. -0.04 ruf. -1.80 dom. +0.13 bre. -0.14 |
HIS | chainID: YYY, resSeq: 381, molecule: Octaheme nitrite reductase |
oxidoreductase oligomeric count: 3 pocket vol.: 519.0 Å3 d(Fe-oop): 0.069 Å |
| HIS | chainID: YYY, resSeq: 424, molecule: Octaheme nitrite reductase |
|||
