Browser
Protein data
function: translocaseexperiment: ELECTRON MICROSCOPY
resolution: 3.1 Å
origomeric count: 22
axial ligand #1: HIS
chainID: N,resSeq: 123,
Coordination distance[Å]: 2.543
molecule: Cytochrome bc1 complex cytochrome b subunit
EC number: 7.1.1.8
Organism: Mycolicibacterium smegmatis MC2 155
axial ligand #2: HIS
chainID: N,resSeq: 226,
Coordination distance[Å]: 2.432
molecule: same as ligand #1
List of other hemes in pdb:9gy6
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
9gy6-B-601 |
sad. -1.30 ruf. -0.41 dom. +0.12 bre. -0.04 |
HIS | chainID: B, resSeq: 123, molecule: Cytochrome bc1 complex cytochrome b subunit |
translocase oligomeric count: 22 pocket vol.: 369.0 Å3 d(Fe-oop): 0.098 Å |
| HIS | chainID: B, resSeq: 226, molecule: Cytochrome bc1 complex cytochrome b subunit |
|||
9gy6-B-602 |
sad. -0.66 ruf. +0.19 dom. -0.01 bre. -0.12 |
HIS | chainID: B, resSeq: 109, molecule: Cytochrome bc1 complex cytochrome b subunit |
translocase oligomeric count: 22 pocket vol.: 375.0 Å3 d(Fe-oop): 0.025 Å |
| HIS | chainID: B, resSeq: 211, molecule: Cytochrome bc1 complex cytochrome b subunit |
|||
9gy6-C-301 |
sad. +0.18 ruf. -0.46 dom. +0.28 bre. -0.22 |
HIS | chainID: C, resSeq: 91, molecule: Cytochrome bc1 complex cytochrome c subunit |
oxidoreductase oligomeric count: 22 pocket vol.: 490.0 Å3 d(Fe-oop): 0.078 Å |
| MET | chainID: C, resSeq: 122, molecule: Cytochrome bc1 complex cytochrome c subunit |
|||
9gy6-C-302 |
sad. +0.19 ruf. -0.75 dom. +0.45 bre. -0.18 |
HIS | chainID: C, resSeq: 192, molecule: Cytochrome bc1 complex cytochrome c subunit |
oxidoreductase oligomeric count: 22 pocket vol.: 360.0 Å3 d(Fe-oop): 0.046 Å |
| MET | chainID: C, resSeq: 229, molecule: Cytochrome bc1 complex cytochrome c subunit |
|||
9gy6-F-601 |
sad. -0.61 ruf. -0.15 dom. +0.50 bre. -0.49 |
HIS | chainID: F, resSeq: 397, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 22 pocket vol.: 746.0 Å3 d(Fe-oop): 0.062 Å |
| Not assigned. | ||||
9gy6-F-602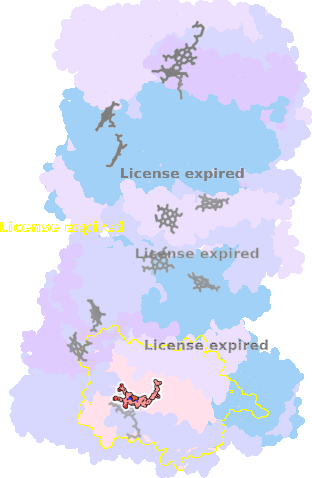 |
sad. -0.74 ruf. +1.64 dom. +0.63 bre. -0.22 |
HIS | chainID: F, resSeq: 86, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 22 pocket vol.: 690.0 Å3 d(Fe-oop): 0.014 Å |
| HIS | chainID: F, resSeq: 399, molecule: Cytochrome c oxidase subunit 1 |
|||
9gy6-N-602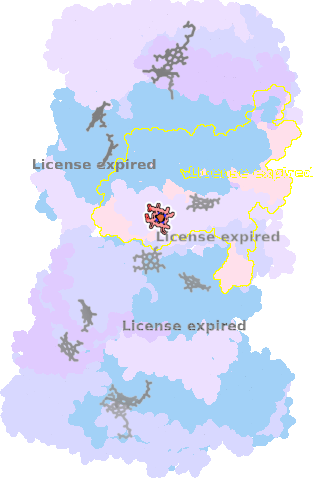 |
sad. -0.66 ruf. -0.01 dom. -0.00 bre. -0.11 |
HIS | chainID: N, resSeq: 109, molecule: Cytochrome bc1 complex cytochrome b subunit |
translocase oligomeric count: 22 pocket vol.: 388.0 Å3 d(Fe-oop): 0.055 Å |
| HIS | chainID: N, resSeq: 211, molecule: Cytochrome bc1 complex cytochrome b subunit |
|||
9gy6-O-302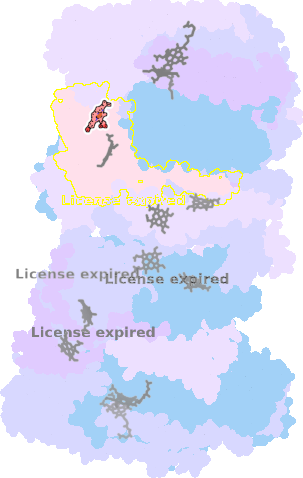 |
sad. +0.39 ruf. -0.40 dom. +0.63 bre. -0.19 |
HIS | chainID: O, resSeq: 91, molecule: Cytochrome bc1 complex cytochrome c subunit |
oxidoreductase oligomeric count: 22 pocket vol.: 430.0 Å3 d(Fe-oop): 0.041 Å |
| MET | chainID: O, resSeq: 122, molecule: Cytochrome bc1 complex cytochrome c subunit |
|||
9gy6-O-303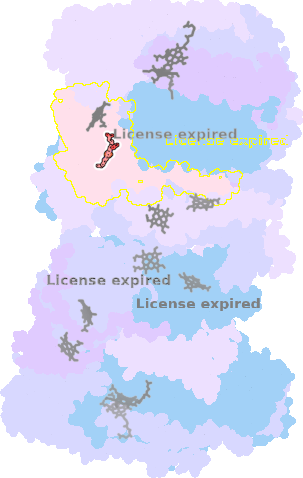 |
sad. +0.12 ruf. -0.95 dom. +0.31 bre. -0.13 |
HIS | chainID: O, resSeq: 192, molecule: Cytochrome bc1 complex cytochrome c subunit |
oxidoreductase oligomeric count: 22 pocket vol.: 353.0 Å3 d(Fe-oop): 0.029 Å |
| MET | chainID: O, resSeq: 229, molecule: Cytochrome bc1 complex cytochrome c subunit |
|||
9gy6-R-601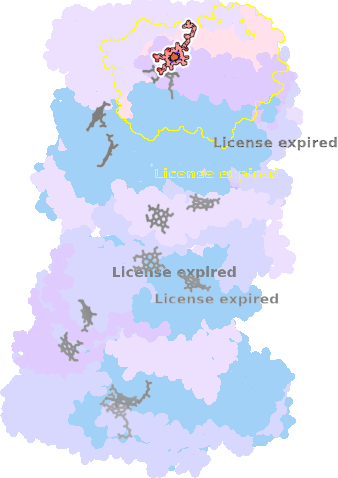 |
sad. -0.70 ruf. +0.15 dom. +0.56 bre. -0.45 |
HIS | chainID: R, resSeq: 397, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 22 pocket vol.: 768.0 Å3 d(Fe-oop): 0.056 Å |
| Not assigned. | ||||
9gy6-R-602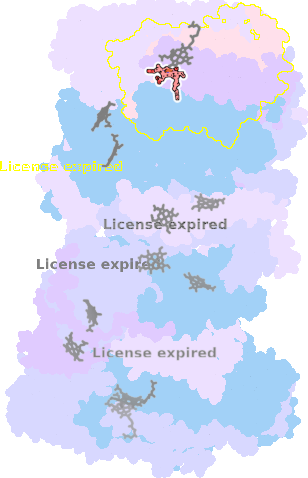 |
sad. -0.68 ruf. +1.27 dom. +0.69 bre. -0.30 |
HIS | chainID: R, resSeq: 86, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 22 pocket vol.: 701.0 Å3 d(Fe-oop): 0.007 Å |
| HIS | chainID: R, resSeq: 399, molecule: Cytochrome c oxidase subunit 1 |
|||
