Browser
Protein data
function: electron transporterexperiment: ELECTRON MICROSCOPY
resolution: 4.03 Å
origomeric count: 31
axial ligand #1: HIS
chainID: b,resSeq: 249,
Coordination distance[Å]: 2.095
molecule: Cytochrome c1
Organism: Paracoccus denitrificans PD1222
axial ligand #2: MET
chainID: b,resSeq: 373,
Coordination distance[Å]: 2.264
molecule: same as ligand #1
List of other hemes in pdb:9g9z
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
9g9z-a-505 |
sad. -0.01 ruf. +0.23 dom. +0.02 bre. +0.13 |
HIS | chainID: a, resSeq: 97, molecule: Cytochrome b |
electron transporter oligomeric count: 31 pocket vol.: 445.0 Å3 d(Fe-oop): 0.080 Å |
| HIS | chainID: a, resSeq: 198, molecule: Cytochrome b |
|||
9g9z-a-507 |
sad. -0.56 ruf. +0.18 dom. +0.18 bre. +0.16 |
HIS | chainID: a, resSeq: 111, molecule: Cytochrome b |
electron transporter oligomeric count: 31 pocket vol.: 454.0 Å3 d(Fe-oop): 0.063 Å |
| HIS | chainID: a, resSeq: 212, molecule: Cytochrome b |
|||
9g9z-d-502 |
sad. -0.41 ruf. -0.89 dom. +0.41 bre. +0.16 |
HIS | chainID: d, resSeq: 111, molecule: Cytochrome b |
electron transporter oligomeric count: 31 pocket vol.: 477.0 Å3 d(Fe-oop): 0.005 Å |
| HIS | chainID: d, resSeq: 212, molecule: Cytochrome b |
|||
9g9z-d-503 |
sad. -0.02 ruf. +0.12 dom. -0.04 bre. +0.06 |
HIS | chainID: d, resSeq: 97, molecule: Cytochrome b |
electron transporter oligomeric count: 31 pocket vol.: 454.0 Å3 d(Fe-oop): 0.068 Å |
| HIS | chainID: d, resSeq: 198, molecule: Cytochrome b |
|||
9g9z-e-502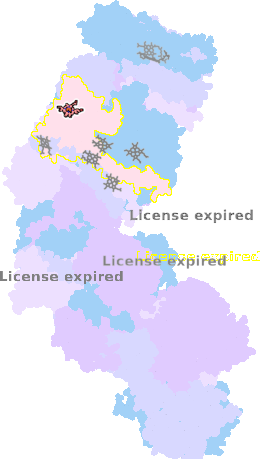 |
sad. -0.15 ruf. -0.27 dom. +0.18 bre. +0.00 |
HIS | chainID: e, resSeq: 249, molecule: Cytochrome c1 |
electron transporter oligomeric count: 31 pocket vol.: 417.0 Å3 d(Fe-oop): 0.096 Å |
| MET | chainID: e, resSeq: 373, molecule: Cytochrome c1 |
|||
9g9z-g-601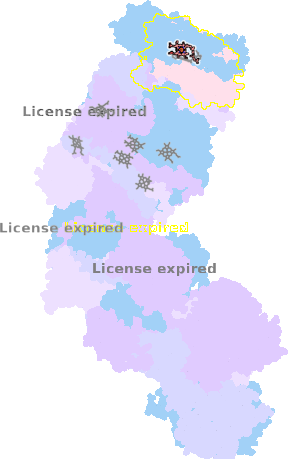 |
sad. -0.02 ruf. +0.01 dom. +0.01 bre. +0.07 |
HIS | chainID: g, resSeq: 94, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 31 pocket vol.: 666.0 Å3 d(Fe-oop): 0.012 Å |
| HIS | chainID: g, resSeq: 413, molecule: Cytochrome c oxidase subunit 1 |
|||
9g9z-g-604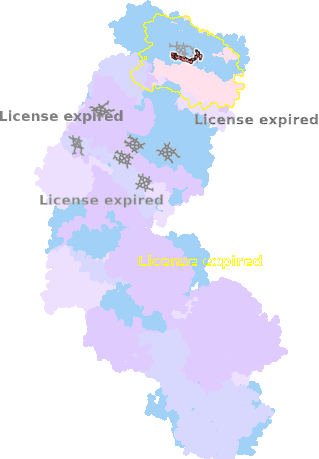 |
sad. +0.01 ruf. +0.01 dom. +0.02 bre. +0.07 |
HIS | chainID: g, resSeq: 411, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 31 pocket vol.: 734.0 Å3 d(Fe-oop): 0.033 Å |
| Not assigned. | ||||
