Browser
Protein data
function: electron transporterexperiment: ELECTRON MICROSCOPY
resolution: 4.91 Å
origomeric count: 54
axial ligand #1: HIS
chainID: d,resSeq: 111,
Coordination distance[Å]: 2.069
molecule: Cytochrome b
Organism: Paracoccus denitrificans PD1222
axial ligand #2: HIS
chainID: d,resSeq: 212,
Coordination distance[Å]: 2.079
molecule: same as ligand #1
List of other hemes in pdb:9g9y
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
9g9y-a-1001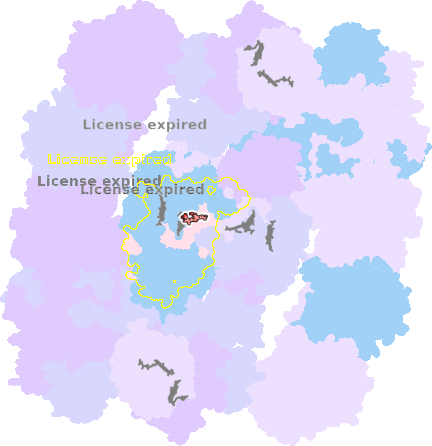 |
sad. -0.02 ruf. +0.35 dom. -0.06 bre. +0.37 |
HIS | chainID: a, resSeq: 97, molecule: Cytochrome b |
electron transporter oligomeric count: 54 pocket vol.: 438.0 Å3 d(Fe-oop): 0.018 Å |
| HIS | chainID: a, resSeq: 198, molecule: Cytochrome b |
|||
9g9y-a-1003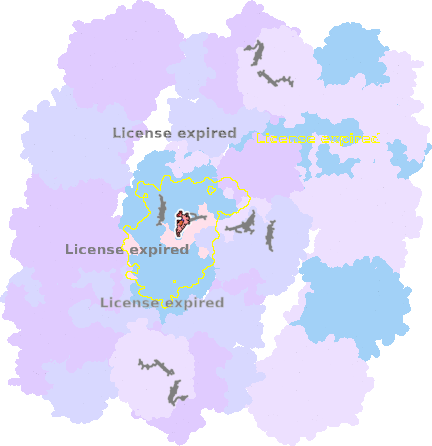 |
sad. -0.28 ruf. -0.80 dom. +0.44 bre. +0.43 |
HIS | chainID: a, resSeq: 111, molecule: Cytochrome b |
electron transporter oligomeric count: 54 pocket vol.: 428.0 Å3 d(Fe-oop): 0.170 Å |
| HIS | chainID: a, resSeq: 212, molecule: Cytochrome b |
|||
9g9y-b-503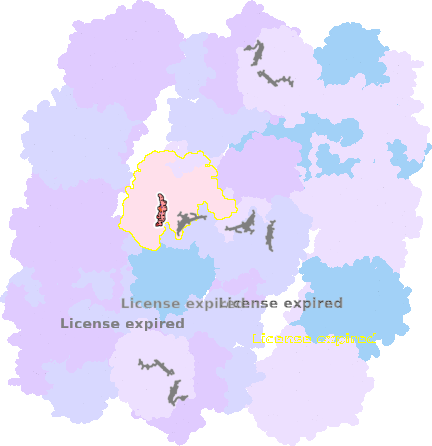 |
sad. -0.06 ruf. +0.06 dom. +0.36 bre. +0.31 |
HIS | chainID: b, resSeq: 249, molecule: Cytochrome c1 |
electron transporter oligomeric count: 54 pocket vol.: 441.0 Å3 d(Fe-oop): 0.026 Å |
| MET | chainID: b, resSeq: 373, molecule: Cytochrome c1 |
|||
9g9y-d-505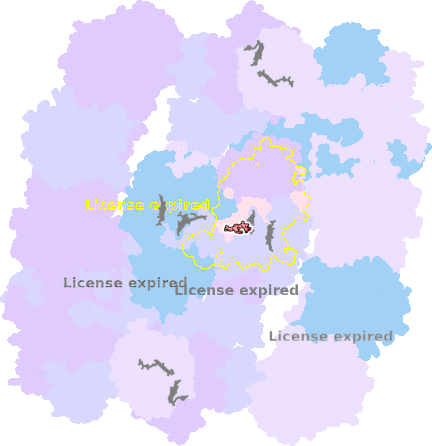 |
sad. +0.00 ruf. +0.29 dom. -0.08 bre. +0.36 |
HIS | chainID: d, resSeq: 97, molecule: Cytochrome b |
electron transporter oligomeric count: 54 pocket vol.: 451.0 Å3 d(Fe-oop): 0.029 Å |
| HIS | chainID: d, resSeq: 198, molecule: Cytochrome b |
|||
9g9y-e-503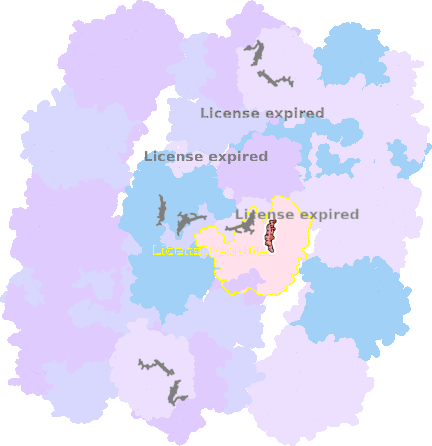 |
sad. -0.06 ruf. +0.06 dom. +0.36 bre. +0.31 |
HIS | chainID: e, resSeq: 249, molecule: Cytochrome c1 |
electron transporter oligomeric count: 54 pocket vol.: 439.0 Å3 d(Fe-oop): 0.026 Å |
| MET | chainID: e, resSeq: 373, molecule: Cytochrome c1 |
|||
9g9y-g-601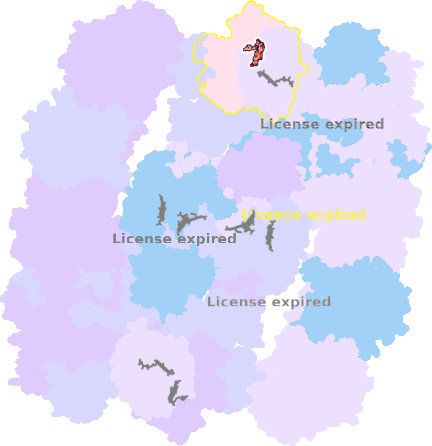 |
sad. -0.00 ruf. +0.01 dom. +0.00 bre. +0.14 |
HIS | chainID: g, resSeq: 94, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 54 pocket vol.: 671.0 Å3 d(Fe-oop): 0.007 Å |
| HIS | chainID: g, resSeq: 413, molecule: Cytochrome c oxidase subunit 1 |
|||
9g9y-g-605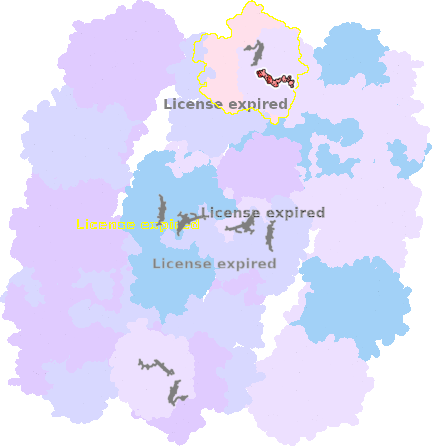 |
sad. -0.00 ruf. +0.01 dom. +0.02 bre. +0.15 |
HIS | chainID: g, resSeq: 411, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 54 pocket vol.: 758.0 Å3 d(Fe-oop): 0.028 Å |
| Not assigned. | ||||
9g9y-k-603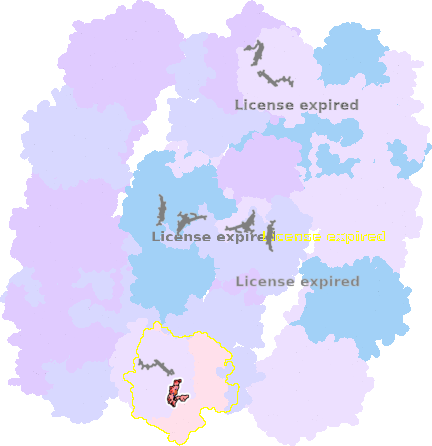 |
sad. -0.00 ruf. +0.01 dom. +0.00 bre. +0.14 |
HIS | chainID: k, resSeq: 94, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 54 pocket vol.: 682.0 Å3 d(Fe-oop): 0.007 Å |
| HIS | chainID: k, resSeq: 413, molecule: Cytochrome c oxidase subunit 1 |
|||
9g9y-k-605 |
sad. -0.00 ruf. +0.01 dom. +0.02 bre. +0.15 |
HIS | chainID: k, resSeq: 411, molecule: Cytochrome c oxidase subunit 1 |
translocase oligomeric count: 54 pocket vol.: 755.0 Å3 d(Fe-oop): 0.030 Å |
| Not assigned. | ||||
