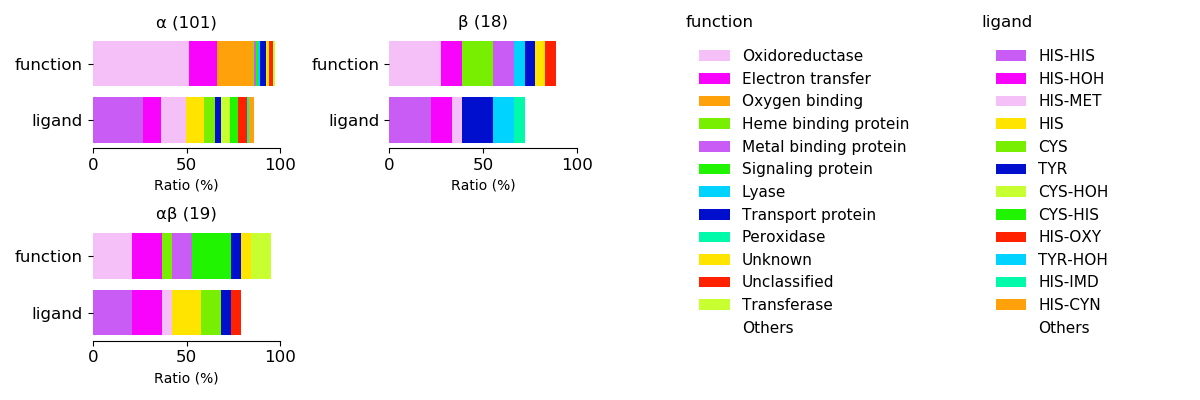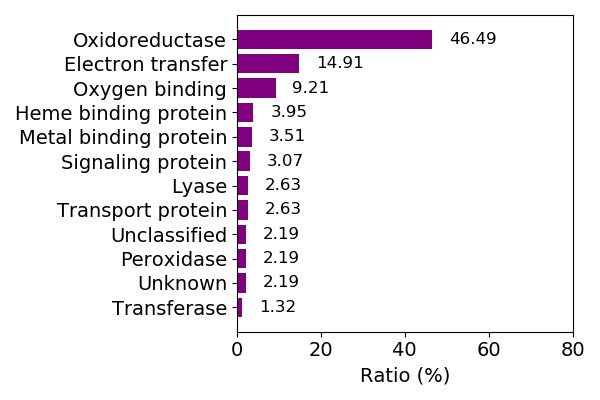Non-redundant dataset
Non-redundant datasets were extracted from whole data in PyDISH by using PISCES (G. Wang and R. L. Dunbrack Jr, Nucleic Acids Res. 33, W94, 2005). The thresholds for sequence similarity were specified to 25% and 40%, and 228 and 423 PDB chains were extracted, respectively. We selected the PyDISH entries including these PDB chains and analyzed the compositions of axial ligands, protein function, and protein fold (CATH level C).
Composition of the axial ligands
Sequence similality ≤ 40%

Sequence similality ≤ 25%

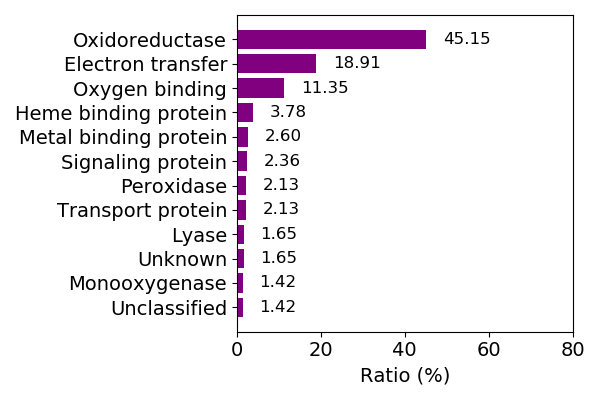
Composition of the protein function
Composition of the protein fold
Sequence similality ≤ 40%
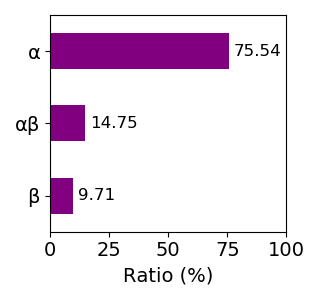
Sequence similality ≤ 25%
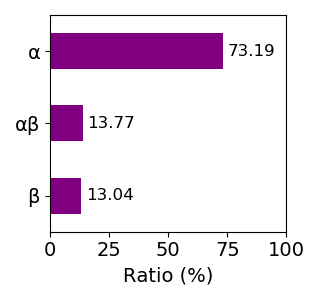
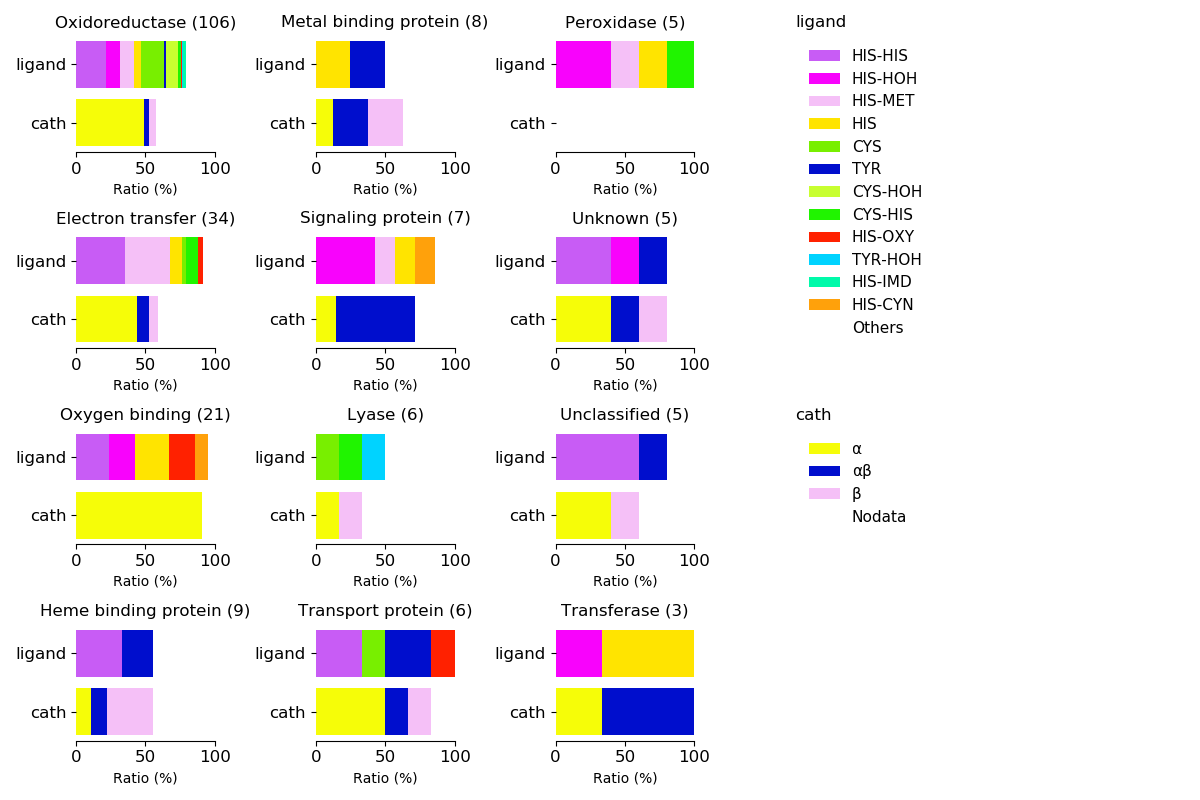
Axial ligands and protein fold for respective functions
Sequence similality ≤ 40%
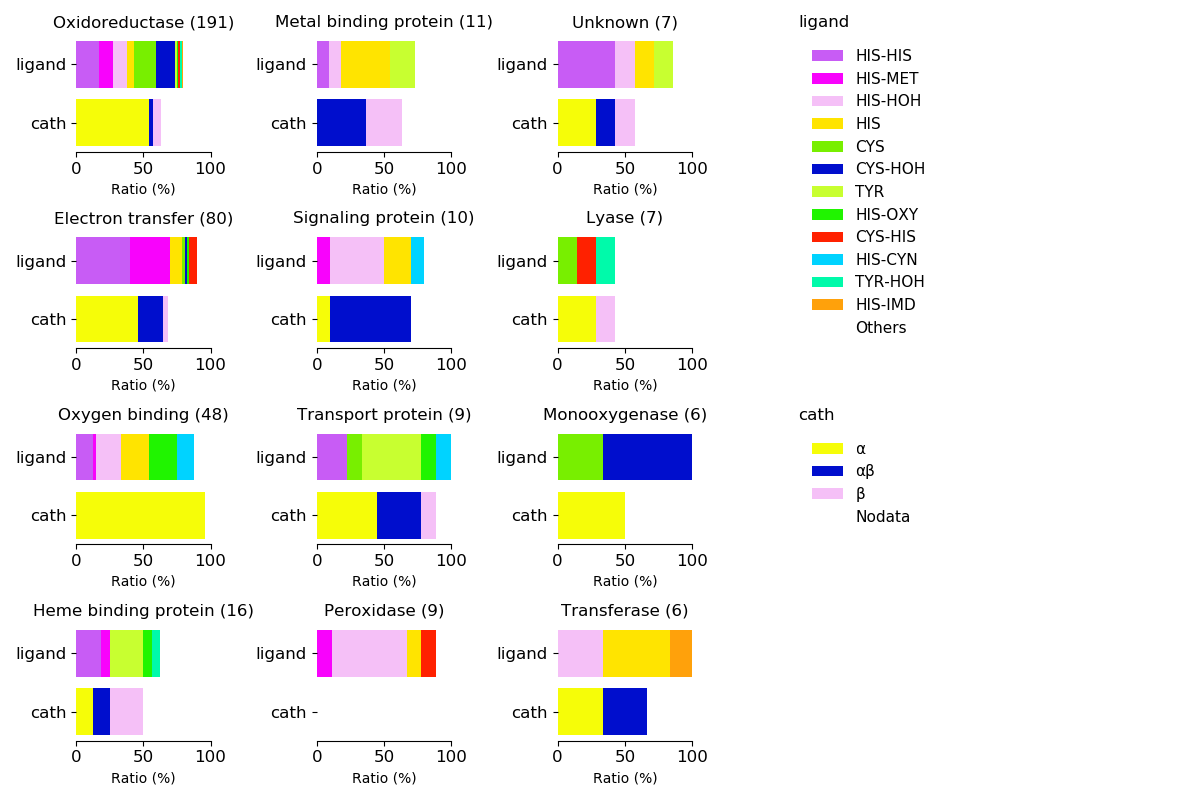
Sequence similality ≤ 25%
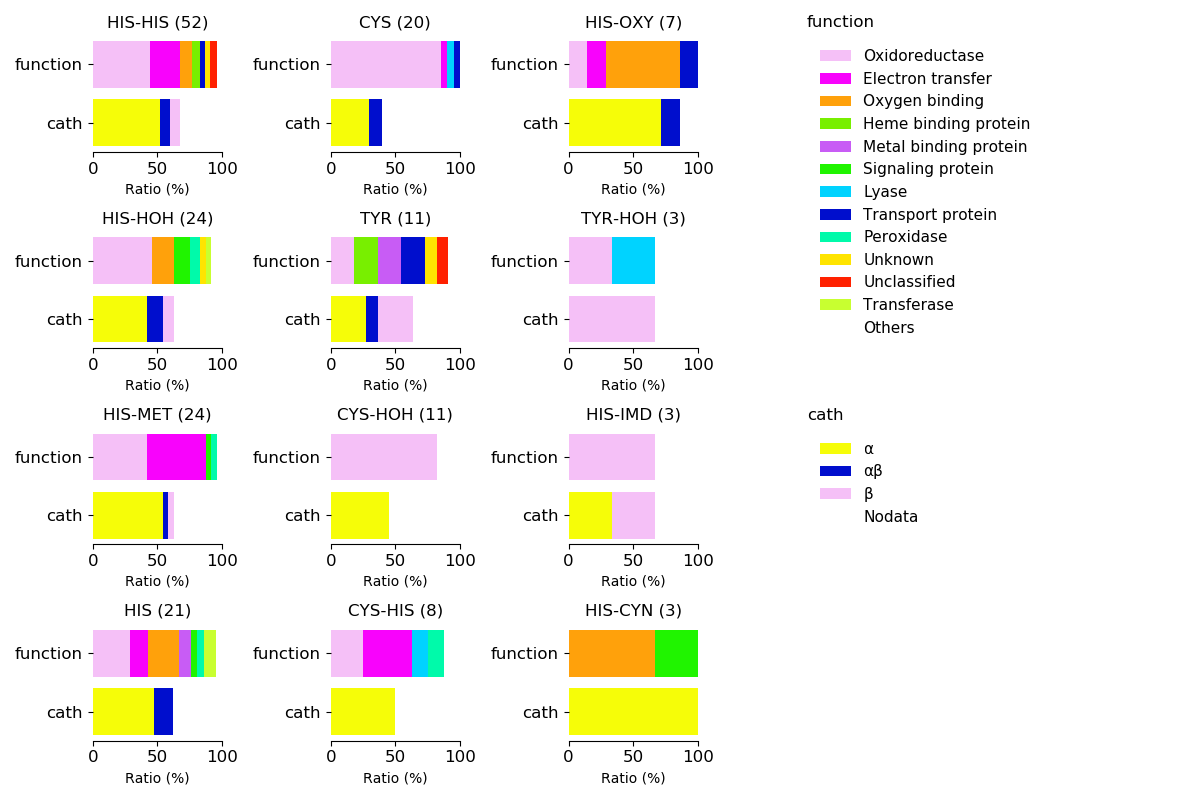
Protein function and protein fold for respective axial ligands
Sequence similality ≤ 40%

Sequence similality ≤ 25%
Axial ligands and protein function for respective protein folds
Sequence similality ≤ 40%
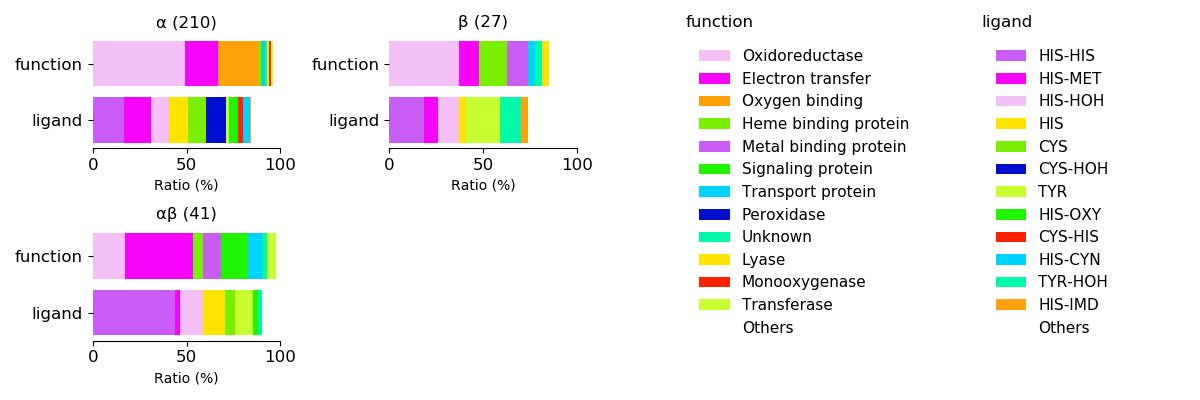
Sequence similality ≤ 25%