Browser
Protein data
function: electron transporterexperiment: ELECTRON MICROSCOPY
resolution: 1.94 Å
origomeric count: 18
axial ligand #1: TYR
chainID: C,resSeq: 1,
Coordination distance[Å]: 2.188
molecule: Cytochrome f
Organism: Spinacia oleracea
axial ligand #2: HIS
chainID: C,resSeq: 25,
Coordination distance[Å]: 2.288
molecule: same as ligand #1
List of other hemes in pdb:9es7
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
9es7-A-301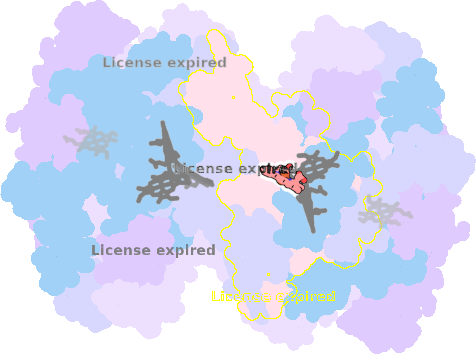 |
sad. -0.08 ruf. +0.03 dom. +0.03 bre. -0.22 |
HIS | chainID: A, resSeq: 86, molecule: Cytochrome b6 |
electron transporter oligomeric count: 18 pocket vol.: 483.0 Å3 d(Fe-oop): 0.015 Å |
| HIS | chainID: A, resSeq: 187, molecule: Cytochrome b6 |
|||
9es7-A-302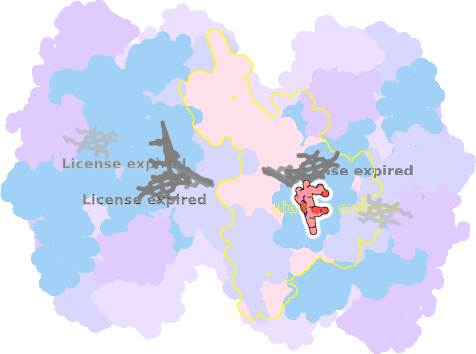 |
sad. -0.84 ruf. -0.44 dom. +0.10 bre. -0.16 |
HIS | chainID: A, resSeq: 100, molecule: Cytochrome b6 |
electron transporter oligomeric count: 18 pocket vol.: 497.0 Å3 d(Fe-oop): 0.012 Å |
| HIS | chainID: A, resSeq: 202, molecule: Cytochrome b6 |
|||
9es7-A-303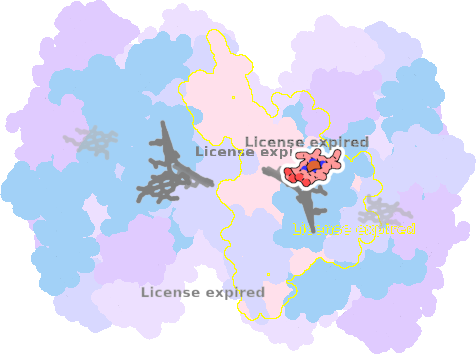 |
sad. +0.56 ruf. -0.04 dom. -0.17 bre. -0.26 |
HOH | chainID: A, resSeq: 405, molecule: water |
electron transporter oligomeric count: 18 pocket vol.: 679.0 Å3 d(Fe-oop): 0.107 Å |
| Not assigned. | ||||
9es7-I-301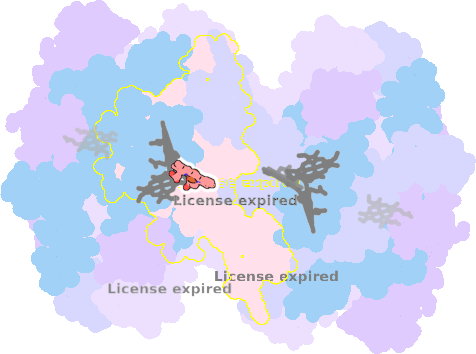 |
sad. -0.08 ruf. +0.03 dom. +0.03 bre. -0.22 |
HIS | chainID: I, resSeq: 86, molecule: Cytochrome b6 |
electron transporter oligomeric count: 18 pocket vol.: 503.0 Å3 d(Fe-oop): 0.016 Å |
| HIS | chainID: I, resSeq: 187, molecule: Cytochrome b6 |
|||
9es7-I-302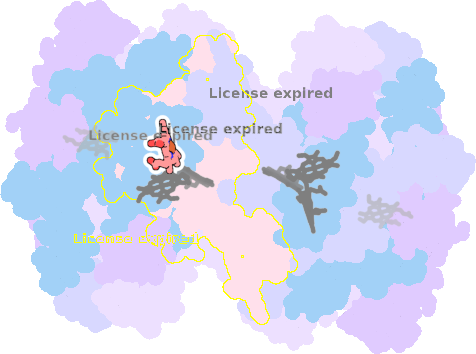 |
sad. -0.84 ruf. -0.44 dom. +0.10 bre. -0.16 |
HIS | chainID: I, resSeq: 100, molecule: Cytochrome b6 |
electron transporter oligomeric count: 18 pocket vol.: 489.0 Å3 d(Fe-oop): 0.012 Å |
| HIS | chainID: I, resSeq: 202, molecule: Cytochrome b6 |
|||
9es7-I-303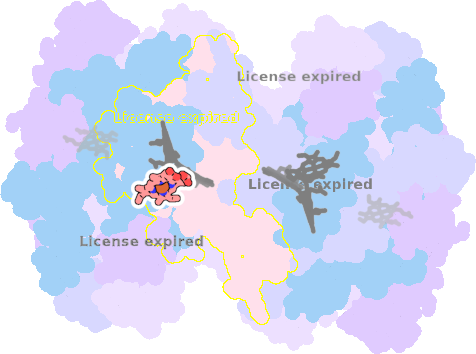 |
sad. +0.56 ruf. -0.04 dom. -0.17 bre. -0.26 |
HOH | chainID: I, resSeq: 405, molecule: water |
electron transporter oligomeric count: 18 pocket vol.: 654.0 Å3 d(Fe-oop): 0.107 Å |
| Not assigned. | ||||
9es7-K-301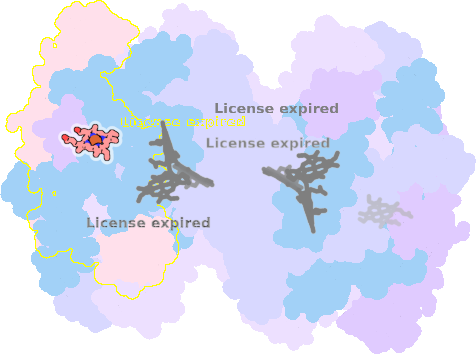 |
sad. -0.20 ruf. -0.68 dom. +0.02 bre. -0.21 |
TYR | chainID: K, resSeq: 1, molecule: Cytochrome f |
electron transporter oligomeric count: 18 pocket vol.: 451.0 Å3 d(Fe-oop): 0.118 Å |
| HIS | chainID: K, resSeq: 25, molecule: Cytochrome f |
|||
