Browser
Protein data
function: electron transporterexperiment: ELECTRON MICROSCOPY
resolution: 2.6 Å
origomeric count: 36
axial ligand #1: HIS
chainID: C,resSeq: 145,
Coordination distance[Å]: 2.328
molecule: Photosynthetic reaction center cytochrome c subunit
Organism: Halorhodospira halophila
axial ligand #2: HIS
chainID: C,resSeq: 316,
Coordination distance[Å]: 1.857
molecule: same as ligand #1
List of other hemes in pdb:8z83
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
8z83-C-401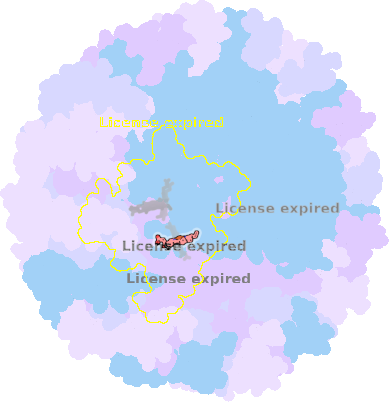 |
sad. +0.21 ruf. -0.21 dom. -0.07 bre. +0.01 |
MET | chainID: C, resSeq: 95, molecule: Photosynthetic reaction center cytochrome c subunit |
electron transporter oligomeric count: 36 pocket vol.: 339.0 Å3 d(Fe-oop): 0.005 Å |
| HIS | chainID: C, resSeq: 112, molecule: Photosynthetic reaction center cytochrome c subunit |
|||
8z83-C-402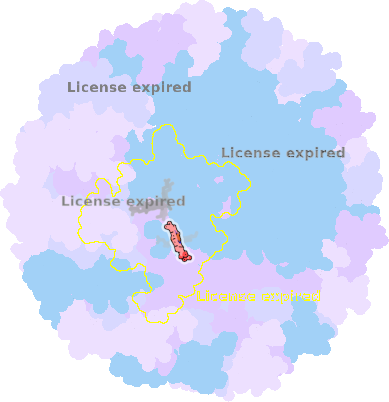 |
sad. -0.08 ruf. -0.53 dom. -0.06 bre. +0.03 |
MET | chainID: C, resSeq: 131, molecule: Photosynthetic reaction center cytochrome c subunit |
electron transporter oligomeric count: 36 pocket vol.: 348.0 Å3 d(Fe-oop): 0.070 Å |
| HIS | chainID: C, resSeq: 157, molecule: Photosynthetic reaction center cytochrome c subunit |
|||
8z83-C-403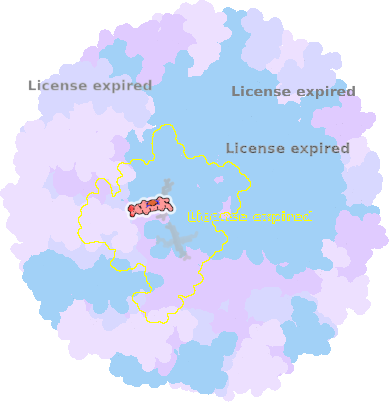 |
sad. -0.02 ruf. -0.29 dom. +0.08 bre. +0.02 |
MET | chainID: C, resSeq: 241, molecule: Photosynthetic reaction center cytochrome c subunit |
electron transporter oligomeric count: 36 pocket vol.: 369.0 Å3 d(Fe-oop): 0.002 Å |
| HIS | chainID: C, resSeq: 256, molecule: Photosynthetic reaction center cytochrome c subunit |
|||
