Browser
Protein data
function: electron transporterexperiment: X-RAY DIFFRACTION
resolution: 2.0 Å
origomeric count: 1
axial ligand #1: HIS
chainID: A,resSeq: 627,
Coordination distance[Å]: 2.28
molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC
Organism: Shewanella oneidensis MR-1
axial ligand #2: HIS
chainID: A,resSeq: 659,
Coordination distance[Å]: 1.98
molecule: same as ligand #1
List of other hemes in pdb:8qc9
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
8qc9-A-701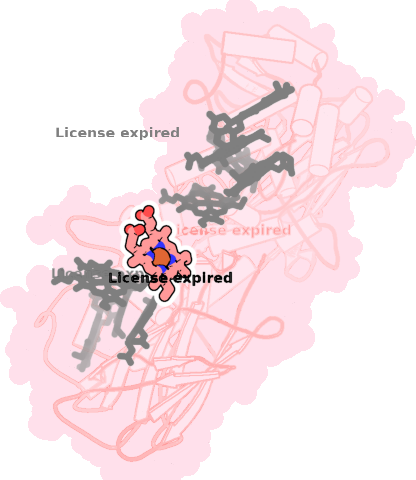 |
sad. -0.30 ruf. -0.46 dom. +0.13 bre. -0.20 |
HIS | chainID: A, resSeq: 190, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
electron transporter oligomeric count: 1 pocket vol.: 453.0 Å3 d(Fe-oop): 0.064 Å |
| HIS | chainID: A, resSeq: 233, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
|||
8qc9-A-702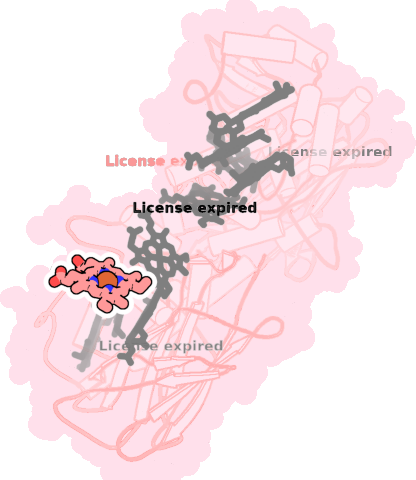 |
sad. +0.23 ruf. -0.06 dom. -0.04 bre. -0.17 |
HIS | chainID: A, resSeq: 198, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
electron transporter oligomeric count: 1 pocket vol.: 437.0 Å3 d(Fe-oop): 0.029 Å |
| HIS | chainID: A, resSeq: 210, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
|||
8qc9-A-703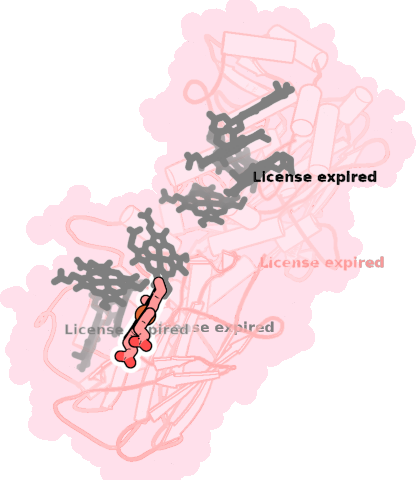 |
sad. -0.21 ruf. -0.18 dom. +0.06 bre. -0.25 |
HIS | chainID: A, resSeq: 230, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
electron transporter oligomeric count: 1 pocket vol.: 410.0 Å3 d(Fe-oop): 0.044 Å |
| HIS | chainID: A, resSeq: 261, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
|||
8qc9-A-704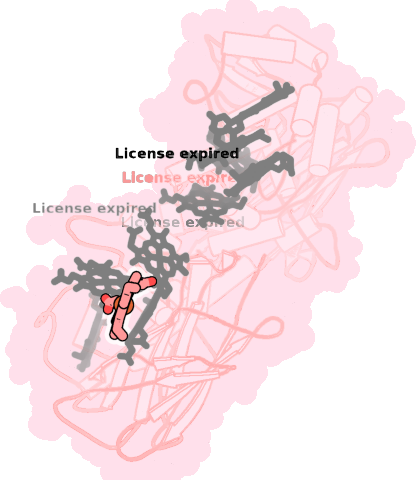 |
sad. +0.48 ruf. -0.17 dom. +0.14 bre. -0.17 |
HIS | chainID: A, resSeq: 286, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
electron transporter oligomeric count: 1 pocket vol.: 599.0 Å3 d(Fe-oop): 0.074 Å |
| HIS | chainID: A, resSeq: 319, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
|||
8qc9-A-705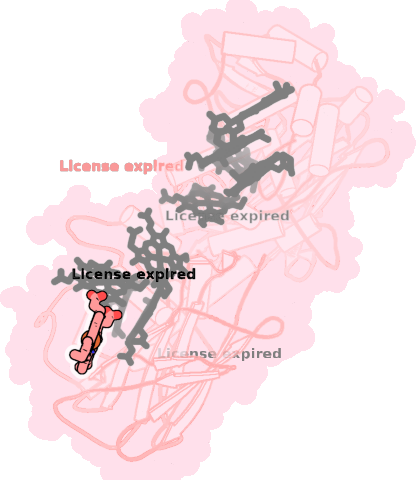 |
sad. +0.05 ruf. -0.63 dom. -0.07 bre. -0.21 |
HIS | chainID: A, resSeq: 297, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
electron transporter oligomeric count: 1 pocket vol.: 595.0 Å3 d(Fe-oop): 0.090 Å |
| HIS | chainID: A, resSeq: 310, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
|||
8qc9-A-706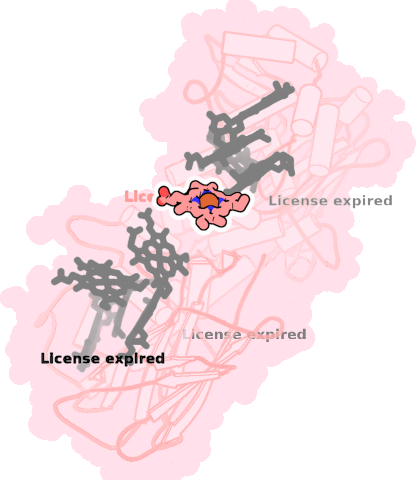 |
sad. +0.41 ruf. -0.59 dom. -0.03 bre. -0.18 |
HIS | chainID: A, resSeq: 500, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
electron transporter oligomeric count: 1 pocket vol.: 504.0 Å3 d(Fe-oop): 0.068 Å |
| HIS | chainID: A, resSeq: 564, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
|||
8qc9-A-707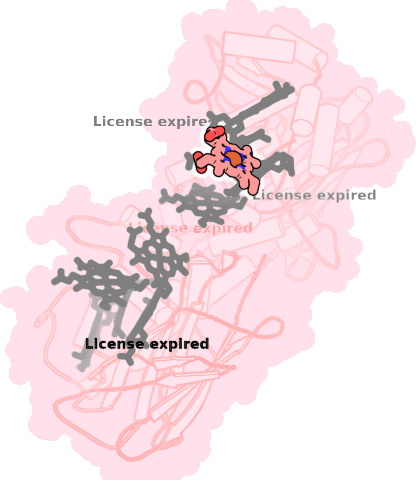 |
sad. -0.06 ruf. -0.21 dom. -0.00 bre. -0.25 |
HIS | chainID: A, resSeq: 507, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
electron transporter oligomeric count: 1 pocket vol.: 467.0 Å3 d(Fe-oop): 0.089 Å |
| HIS | chainID: A, resSeq: 542, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
|||
8qc9-A-708 |
sad. +0.32 ruf. -0.02 dom. -0.03 bre. -0.20 |
HIS | chainID: A, resSeq: 561, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
electron transporter oligomeric count: 1 pocket vol.: 442.0 Å3 d(Fe-oop): 0.044 Å |
| HIS | chainID: A, resSeq: 580, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
|||
8qc9-A-709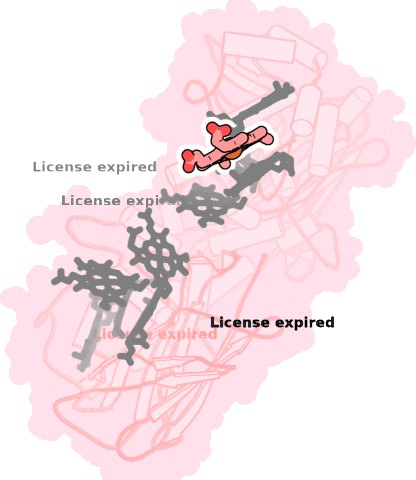 |
sad. +0.07 ruf. +0.06 dom. -0.01 bre. -0.25 |
HIS | chainID: A, resSeq: 618, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
electron transporter oligomeric count: 1 pocket vol.: 630.0 Å3 d(Fe-oop): 0.004 Å |
| HIS | chainID: A, resSeq: 666, molecule: Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC |
|||
