Browser
Protein data
function: oxidoreductaseexperiment: ELECTRON MICROSCOPY
resolution: 3.27 Å
origomeric count: 24
axial ligand #1: MET
chainID: V,resSeq: 52,
Coordination distance[Å]: 1.792
molecule: Bacterioferritin
EC number: 1.16.3.1
Organism: Streptomyces coelicolor
axial ligand #2: MET
chainID: W,resSeq: 52,
Coordination distance[Å]: 2.861
molecule: Bacterioferritin
EC number: 1.16.3.1
Organism: Streptomyces coelicolor
List of other hemes in pdb:8jax
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
8jax-B-201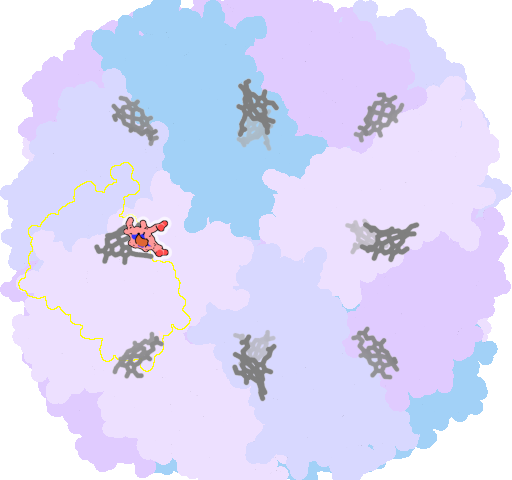 |
sad. -0.47 ruf. -0.91 dom. -0.76 bre. -0.00 |
MET | chainID: A, resSeq: 52, molecule: Bacterioferritin |
oxidoreductase oligomeric count: 24 pocket vol.: 682.0 Å3 d(Fe-oop): 0.242 Å |
| MET | chainID: B, resSeq: 52, molecule: Bacterioferritin |
|||
8jax-D-201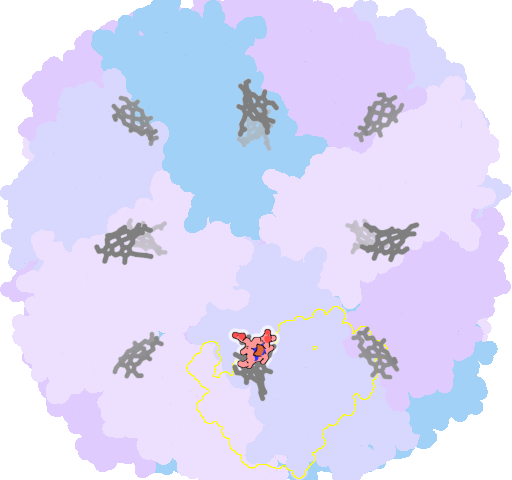 |
sad. -0.54 ruf. +0.53 dom. +0.20 bre. +0.05 |
MET | chainID: C, resSeq: 52, molecule: Bacterioferritin |
oxidoreductase oligomeric count: 24 pocket vol.: 673.0 Å3 d(Fe-oop): 0.150 Å |
| MET | chainID: D, resSeq: 52, molecule: Bacterioferritin |
|||
8jax-F-202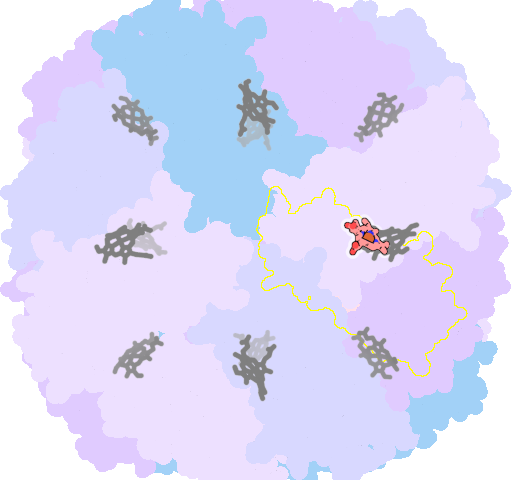 |
sad. +0.02 ruf. -0.79 dom. -0.98 bre. +0.07 |
MET | chainID: F, resSeq: 52, molecule: Bacterioferritin |
oxidoreductase oligomeric count: 24 pocket vol.: 618.0 Å3 d(Fe-oop): 0.152 Å |
| MET | chainID: E, resSeq: 52, molecule: Bacterioferritin |
|||
8jax-H-202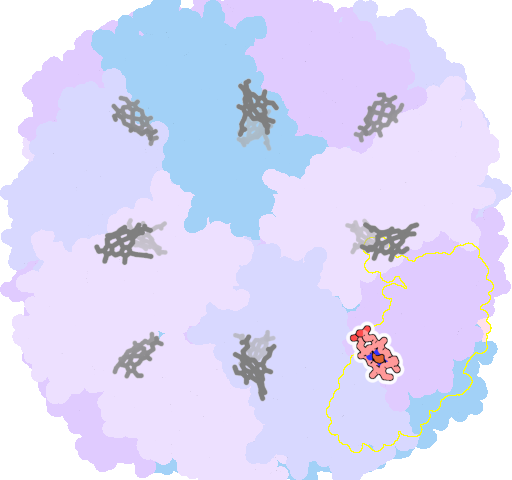 |
sad. -0.38 ruf. -0.62 dom. -0.83 bre. +0.30 |
MET | chainID: H, resSeq: 52, molecule: Bacterioferritin |
oxidoreductase oligomeric count: 24 pocket vol.: 636.0 Å3 d(Fe-oop): 0.429 Å |
| MET | chainID: N, resSeq: 52, molecule: Bacterioferritin |
|||
8jax-K-201 |
sad. -0.57 ruf. +0.30 dom. +1.55 bre. +0.30 |
MET | chainID: J, resSeq: 52, molecule: Bacterioferritin |
oxidoreductase oligomeric count: 24 pocket vol.: 715.0 Å3 d(Fe-oop): 0.315 Å |
| MET | chainID: K, resSeq: 52, molecule: Bacterioferritin |
|||
8jax-M-202 |
sad. -0.79 ruf. -1.09 dom. -0.45 bre. +0.12 |
MET | chainID: L, resSeq: 52, molecule: Bacterioferritin |
oxidoreductase oligomeric count: 24 pocket vol.: 630.0 Å3 d(Fe-oop): 0.264 Å |
| MET | chainID: M, resSeq: 52, molecule: Bacterioferritin |
|||
8jax-P-202 |
sad. -0.50 ruf. -0.58 dom. -0.32 bre. -0.06 |
MET | chainID: O, resSeq: 52, molecule: Bacterioferritin |
oxidoreductase oligomeric count: 24 pocket vol.: 704.0 Å3 d(Fe-oop): 0.173 Å |
| Not assigned. | ||||
8jax-R-201 |
sad. -0.41 ruf. +0.02 dom. -1.30 bre. -0.04 |
MET | chainID: Q, resSeq: 52, molecule: Bacterioferritin |
oxidoreductase oligomeric count: 24 pocket vol.: 679.0 Å3 d(Fe-oop): 0.461 Å |
| MET | chainID: R, resSeq: 52, molecule: Bacterioferritin |
|||
8jax-S-202 |
sad. +0.23 ruf. +0.09 dom. -0.62 bre. +0.16 |
MET | chainID: I, resSeq: 52, molecule: Bacterioferritin |
oxidoreductase oligomeric count: 24 pocket vol.: 592.0 Å3 d(Fe-oop): 0.098 Å |
| MET | chainID: S, resSeq: 52, molecule: Bacterioferritin |
|||
8jax-U-202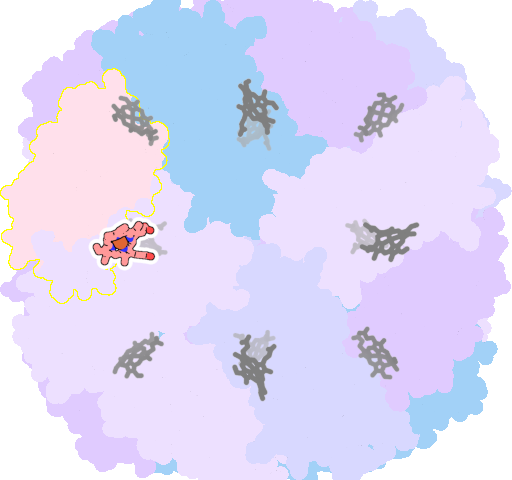 |
sad. -0.54 ruf. +1.01 dom. +1.01 bre. +0.12 |
MET | chainID: T, resSeq: 52, molecule: Bacterioferritin |
oxidoreductase oligomeric count: 24 pocket vol.: 714.0 Å3 d(Fe-oop): 0.115 Å |
| MET | chainID: U, resSeq: 52, molecule: Bacterioferritin |
|||
8jax-X-201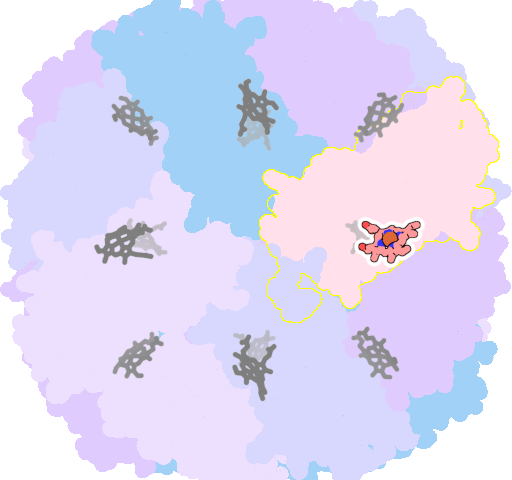 |
sad. -0.12 ruf. +1.36 dom. +0.92 bre. +0.19 |
MET | chainID: G, resSeq: 52, molecule: Bacterioferritin |
oxidoreductase oligomeric count: 24 pocket vol.: 651.0 Å3 d(Fe-oop): 0.222 Å |
| MET | chainID: X, resSeq: 52, molecule: Bacterioferritin |
|||
