Browser
Protein data
function: oxidoreductaseexperiment: ELECTRON MICROSCOPY
resolution: 2.2 Å
origomeric count: 2
axial ligand #1: HIS
chainID: A,resSeq: 331,
Coordination distance[Å]: 1.902
molecule: Nitric oxide reductase subunit B
EC number: 1.7.2.5
Organism: Achromobacter xylosoxidans
axial ligand #2: HIS
chainID: A,resSeq: 631,
Coordination distance[Å]: 1.945
molecule: same as ligand #1
List of other hemes in pdb:8bgw
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
8bgw-A-802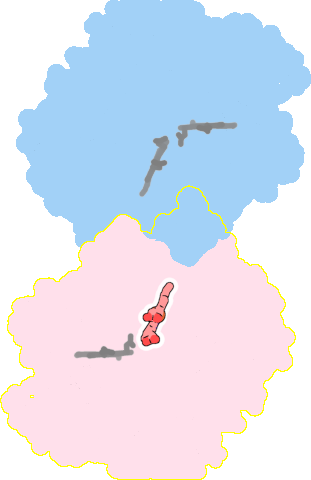 |
sad. -0.43 ruf. -0.05 dom. +0.10 bre. +0.21 |
HIS | chainID: A, resSeq: 629, molecule: Nitric oxide reductase subunit B |
oxidoreductase oligomeric count: 2 pocket vol.: 313.0 Å3 d(Fe-oop): 0.116 Å |
| HOH | chainID: A, resSeq: 935, molecule: water |
|||
8bgw-B-802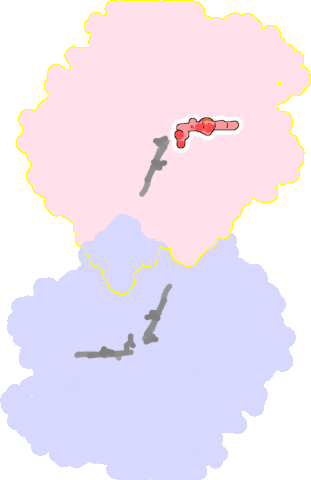 |
sad. +0.31 ruf. -0.13 dom. +0.03 bre. +0.22 |
HIS | chainID: B, resSeq: 331, molecule: Nitric oxide reductase subunit B |
oxidoreductase oligomeric count: 2 pocket vol.: 315.0 Å3 d(Fe-oop): 0.022 Å |
| HIS | chainID: B, resSeq: 631, molecule: Nitric oxide reductase subunit B |
|||
8bgw-B-803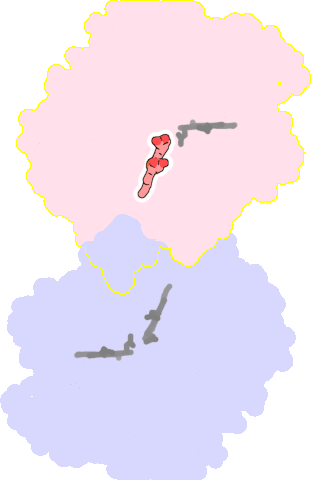 |
sad. -0.43 ruf. -0.06 dom. +0.11 bre. +0.22 |
HIS | chainID: B, resSeq: 629, molecule: Nitric oxide reductase subunit B |
oxidoreductase oligomeric count: 2 pocket vol.: 319.0 Å3 d(Fe-oop): 0.118 Å |
| HOH | chainID: B, resSeq: 930, molecule: water |
|||
