Browser
Protein data
function: unclassifiedexperiment: ELECTRON MICROSCOPY
resolution: 3.0 Å
origomeric count: 6
This entry has no axial ligands.
List of other hemes in pdb:7ose
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
7ose-A-601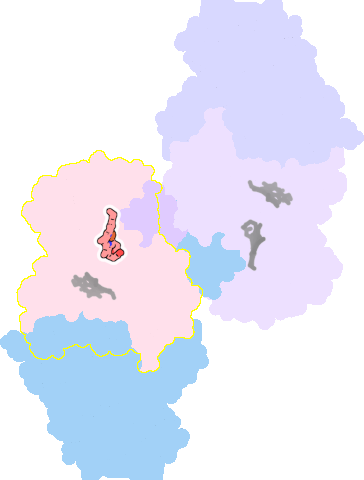 |
sad. +0.00 ruf. +1.85 dom. -0.02 bre. -0.28 |
HIS | chainID: A, resSeq: 186, molecule: Cytochrome bd-II ubiquinol oxidase subunit 1 |
translocase oligomeric count: 6 pocket vol.: 434.0 Å3 d(Fe-oop): 0.012 Å |
| MET | chainID: A, resSeq: 393, molecule: Cytochrome bd-II ubiquinol oxidase subunit 1 |
|||
7ose-A-602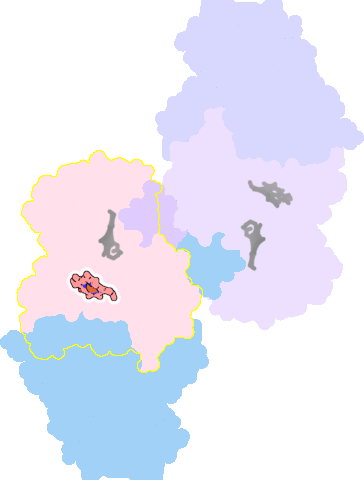 |
sad. +0.09 ruf. -1.78 dom. +0.05 bre. -0.28 |
Not assigned. | unclassified oligomeric count: 6 pocket vol.: 461.0 Å3 d(Fe-oop): 0.003 Å |
|
| Not assigned. | ||||
7ose-D-601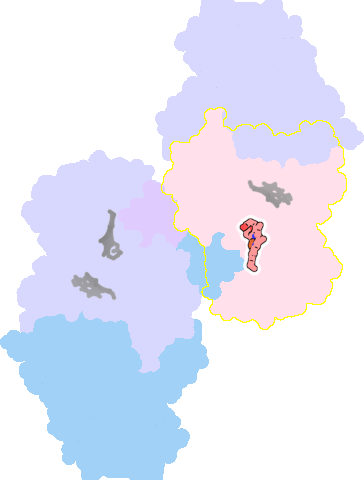 |
sad. +0.00 ruf. +1.85 dom. -0.02 bre. -0.28 |
HIS | chainID: D, resSeq: 186, molecule: Cytochrome bd-II ubiquinol oxidase subunit 1 |
translocase oligomeric count: 6 pocket vol.: 427.0 Å3 d(Fe-oop): 0.012 Å |
| MET | chainID: D, resSeq: 393, molecule: Cytochrome bd-II ubiquinol oxidase subunit 1 |
|||
