Browser
Protein data
function: oxidoreductaseexperiment: X-RAY DIFFRACTION
resolution: 2.6 Å
origomeric count: 5
axial ligand #1: HIS
chainID: B,resSeq: 160,
Coordination distance[Å]: 2.213
molecule: Chlorite dismutase
EC number: 1.13.11.49
Organism: Candidatus Nitrospira defluvii
axial ligand #2: CYN
chainID: B,resSeq: 302,
Coordination distance[Å]: 2.16
molecule: CYANIDE ION
List of other hemes in pdb:4m06
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
4m06-A-301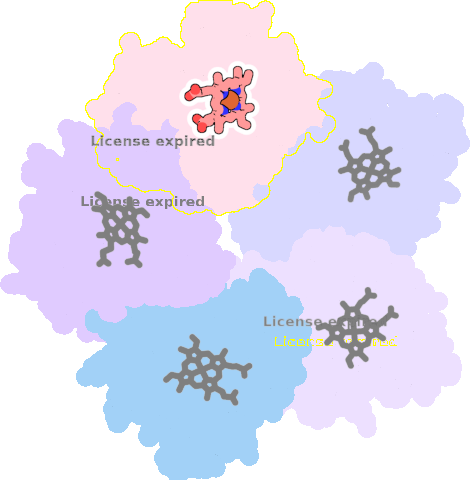 |
sad. -0.38 ruf. +0.40 dom. -0.13 bre. -0.23 |
HIS | chainID: A, resSeq: 160, molecule: Chlorite dismutase |
oxidoreductase oligomeric count: 5 pocket vol.: 462.0 Å3 d(Fe-oop): 0.006 Å |
| CYN | chainID: A, resSeq: 303, molecule: CYANIDE ION |
|||
4m06-C-301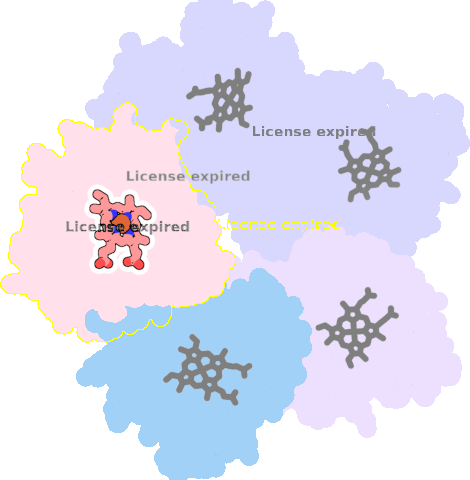 |
sad. -0.43 ruf. +0.41 dom. -0.11 bre. -0.23 |
HIS | chainID: C, resSeq: 160, molecule: Chlorite dismutase |
oxidoreductase oligomeric count: 5 pocket vol.: 464.0 Å3 d(Fe-oop): 0.062 Å |
| CYN | chainID: C, resSeq: 302, molecule: CYANIDE ION |
|||
4m06-D-301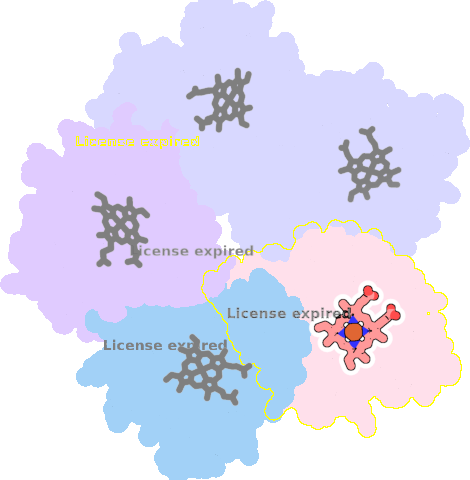 |
sad. -0.44 ruf. +0.56 dom. -0.01 bre. -0.22 |
HIS | chainID: D, resSeq: 160, molecule: Chlorite dismutase |
oxidoreductase oligomeric count: 5 pocket vol.: 465.0 Å3 d(Fe-oop): 0.055 Å |
| CYN | chainID: D, resSeq: 302, molecule: CYANIDE ION |
|||
4m06-E-301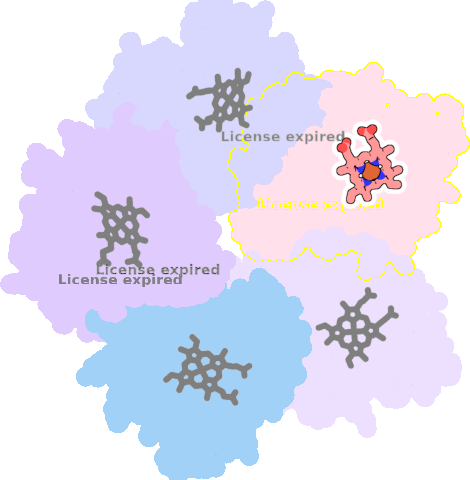 |
sad. -0.43 ruf. +0.46 dom. -0.14 bre. -0.25 |
HIS | chainID: E, resSeq: 160, molecule: Chlorite dismutase |
oxidoreductase oligomeric count: 5 pocket vol.: 472.0 Å3 d(Fe-oop): 0.029 Å |
| CYN | chainID: E, resSeq: 302, molecule: CYANIDE ION |
|||
