Browser
Protein data
function: oxidoreductaseexperiment: X-RAY DIFFRACTION
resolution: 2.399 Å
origomeric count: 1
axial ligand #1: CYS
chainID: D,resSeq: 442,
Coordination distance[Å]: 2.121
molecule: Cytochrome P450 3A4
EC number: 1.14.13.-, 1.14.13.157, 1.14.13.32, 1.14.13.67, 1.14.13.97
Organism: Homo sapiens
axial ligand #2: 7AW
chainID: D,resSeq: 602,
Coordination distance[Å]: 2.314
molecule: N~2~-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-N-[(2R,5S)-1-phenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}octan-2-yl]-L-valinamide
List of other hemes in pdb:4k9w
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
4k9w-A-601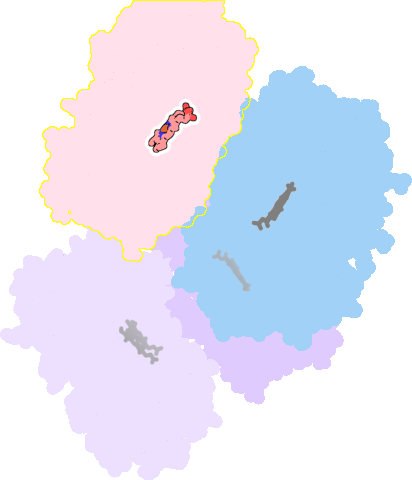 |
sad. -0.31 ruf. +0.43 dom. -0.02 bre. -0.38 |
CYS | chainID: A, resSeq: 442, molecule: Cytochrome P450 3A4 |
oxidoreductase oligomeric count: 1 pocket vol.: 394.0 Å3 d(Fe-oop): 0.378 Å |
| 7AW | chainID: A, resSeq: 602, molecule: N~2~-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-N-[(2R,5S)-1-phenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}octan-2-yl]-L-valinamide |
|||
4k9w-B-601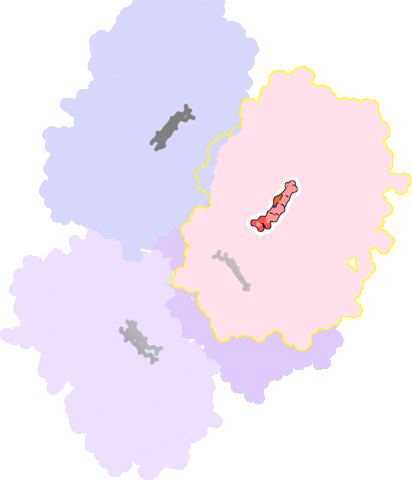 |
sad. -0.39 ruf. +0.37 dom. -0.03 bre. -0.34 |
CYS | chainID: B, resSeq: 442, molecule: Cytochrome P450 3A4 |
oxidoreductase oligomeric count: 1 pocket vol.: 418.0 Å3 d(Fe-oop): 0.428 Å |
| 7AW | chainID: B, resSeq: 602, molecule: N~2~-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-N-[(2R,5S)-1-phenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}octan-2-yl]-L-valinamide |
|||
4k9w-C-601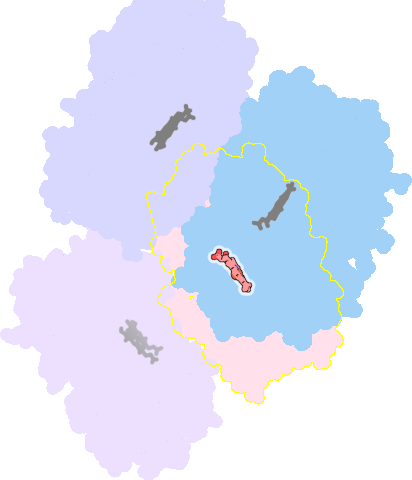 |
sad. -0.46 ruf. +0.37 dom. -0.09 bre. -0.40 |
CYS | chainID: C, resSeq: 442, molecule: Cytochrome P450 3A4 |
oxidoreductase oligomeric count: 1 pocket vol.: 411.0 Å3 d(Fe-oop): 0.427 Å |
| 7AW | chainID: C, resSeq: 602, molecule: N~2~-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-N-[(2R,5S)-1-phenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}octan-2-yl]-L-valinamide |
|||
