Browser
Protein data
function: oxidoreductaseexperiment: X-RAY DIFFRACTION
resolution: 2.0 Å
origomeric count: 1
axial ligand #1: CYS
chainID: D,resSeq: 347,
Coordination distance[Å]: 2.368
molecule: Vitamin D hydroxylase
Organism: Pseudonocardia autotrophica
axial ligand #2: VDY
chainID: D,resSeq: 6178,
Coordination distance[Å]: 2.576
molecule: 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL
List of other hemes in pdb:3a51
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
3a51-A-412 |
sad. -0.20 ruf. -0.48 dom. -0.03 bre. -0.37 |
CYS | chainID: A, resSeq: 347, molecule: Vitamin D hydroxylase |
oxidoreductase oligomeric count: 1 pocket vol.: 398.0 Å3 d(Fe-oop): 0.190 Å |
| VDY | chainID: A, resSeq: 6178, molecule: 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL |
|||
3a51-B-412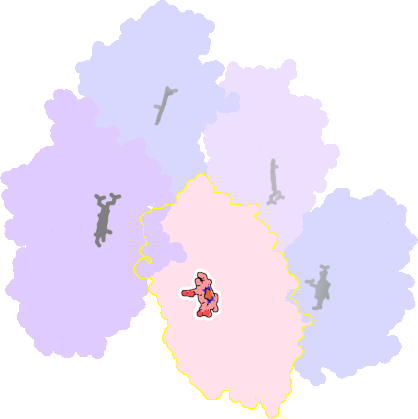 |
sad. -0.17 ruf. -0.41 dom. -0.01 bre. -0.37 |
CYS | chainID: B, resSeq: 347, molecule: Vitamin D hydroxylase |
oxidoreductase oligomeric count: 1 pocket vol.: 395.0 Å3 d(Fe-oop): 0.177 Å |
| VDY | chainID: B, resSeq: 6178, molecule: 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL |
|||
3a51-C-412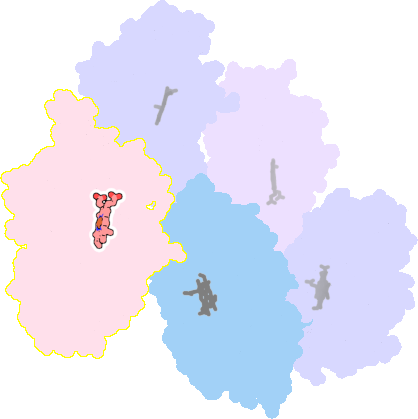 |
sad. -0.28 ruf. -0.39 dom. -0.11 bre. -0.23 |
CYS | chainID: C, resSeq: 347, molecule: Vitamin D hydroxylase |
oxidoreductase oligomeric count: 1 pocket vol.: 405.0 Å3 d(Fe-oop): 0.141 Å |
| VDY | chainID: C, resSeq: 6178, molecule: 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL |
|||
3a51-E-412 |
sad. -0.25 ruf. -0.41 dom. -0.09 bre. -0.29 |
CYS | chainID: E, resSeq: 347, molecule: Vitamin D hydroxylase |
oxidoreductase oligomeric count: 1 pocket vol.: 388.0 Å3 d(Fe-oop): 0.184 Å |
| VDY | chainID: E, resSeq: 6178, molecule: 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL |
|||
