Browser
Protein data
function: electron transporterexperiment: SOLUTION NMR_THEORETICAL MODEL_HYBRID
origomeric count: 3
axial ligand #1: HIS
chainID: E,resSeq: 70,
Coordination distance[Å]: 2.214
molecule: CYTOCHROME C3
Organism: DESULFOVIBRIO VULGARIS
axial ligand #2: HIS
chainID: E,resSeq: 106,
Coordination distance[Å]: 2.279
molecule: same as ligand #1
List of other hemes in pdb:1gx7
| ID of heme | Distortion | Axial ligands on heme | Function & structure | |
|---|---|---|---|---|
1gx7-E-110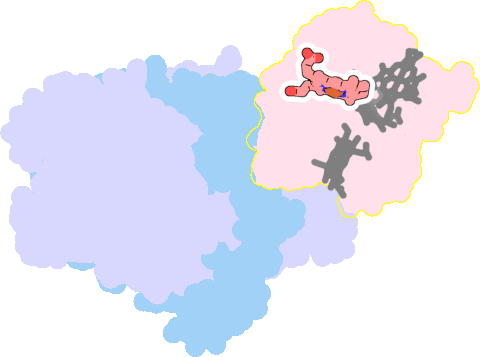 |
sad. -0.02 ruf. -0.68 dom. +0.04 bre. +0.06 |
HIS | chainID: E, resSeq: 35, molecule: CYTOCHROME C3 |
electron transporter oligomeric count: 3 pocket vol.: 439.0 Å3 d(Fe-oop): 0.007 Å |
| HIS | chainID: E, resSeq: 52, molecule: CYTOCHROME C3 |
|||
1gx7-E-111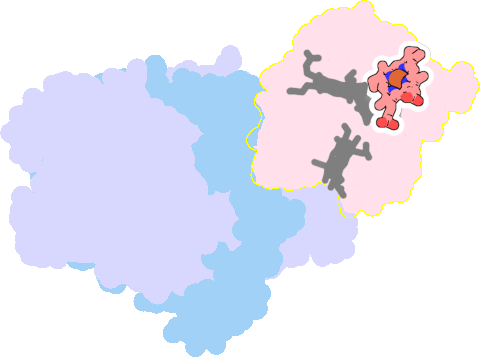 |
sad. +0.58 ruf. -0.41 dom. -0.13 bre. +0.03 |
HIS | chainID: E, resSeq: 22, molecule: CYTOCHROME C3 |
electron transporter oligomeric count: 3 pocket vol.: 440.0 Å3 d(Fe-oop): 0.035 Å |
| HIS | chainID: E, resSeq: 34, molecule: CYTOCHROME C3 |
|||
1gx7-E-112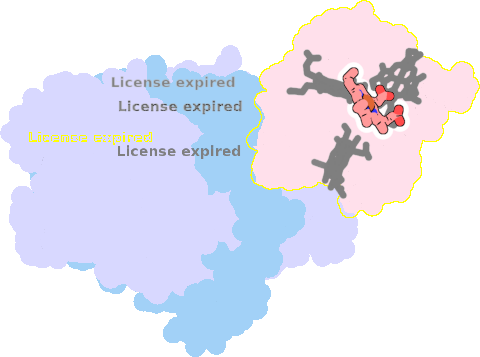 |
sad. -0.38 ruf. -0.41 dom. +0.09 bre. +0.02 |
HIS | chainID: E, resSeq: 25, molecule: CYTOCHROME C3 |
electron transporter oligomeric count: 3 pocket vol.: 504.0 Å3 d(Fe-oop): 0.017 Å |
| HIS | chainID: E, resSeq: 83, molecule: CYTOCHROME C3 |
|||
